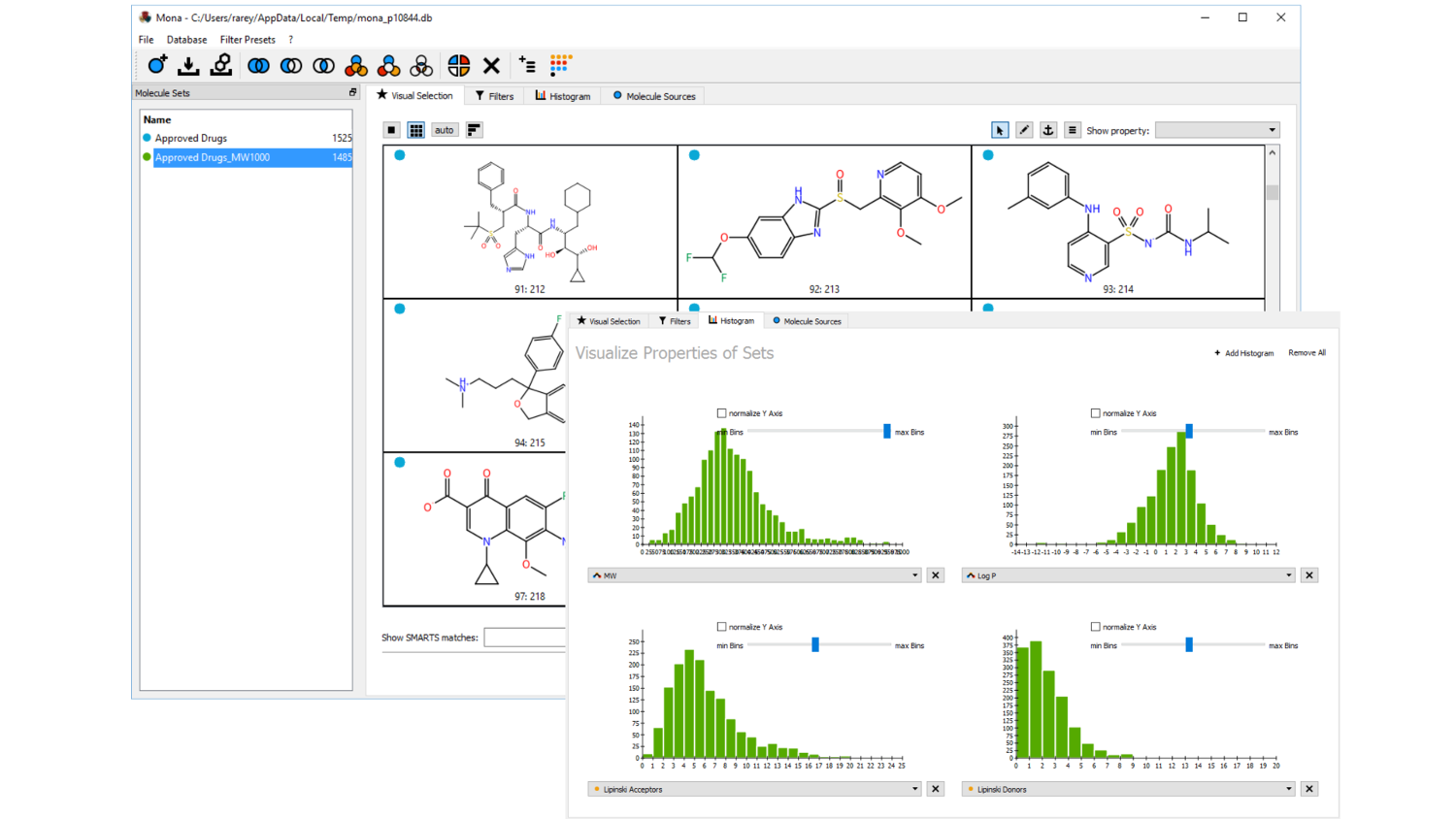
Mona: Managing compound collections
Cheminformatics in an interactive fashion: Visualize, analyze, filter, and cluster compound collections; easily spot the differences between molecule sets; create your own sub-collections by properties, by inclusion and exclusion rules, or just manually. No database installation, no scripting, no pipelining, just an easy-to-use graphical interface.

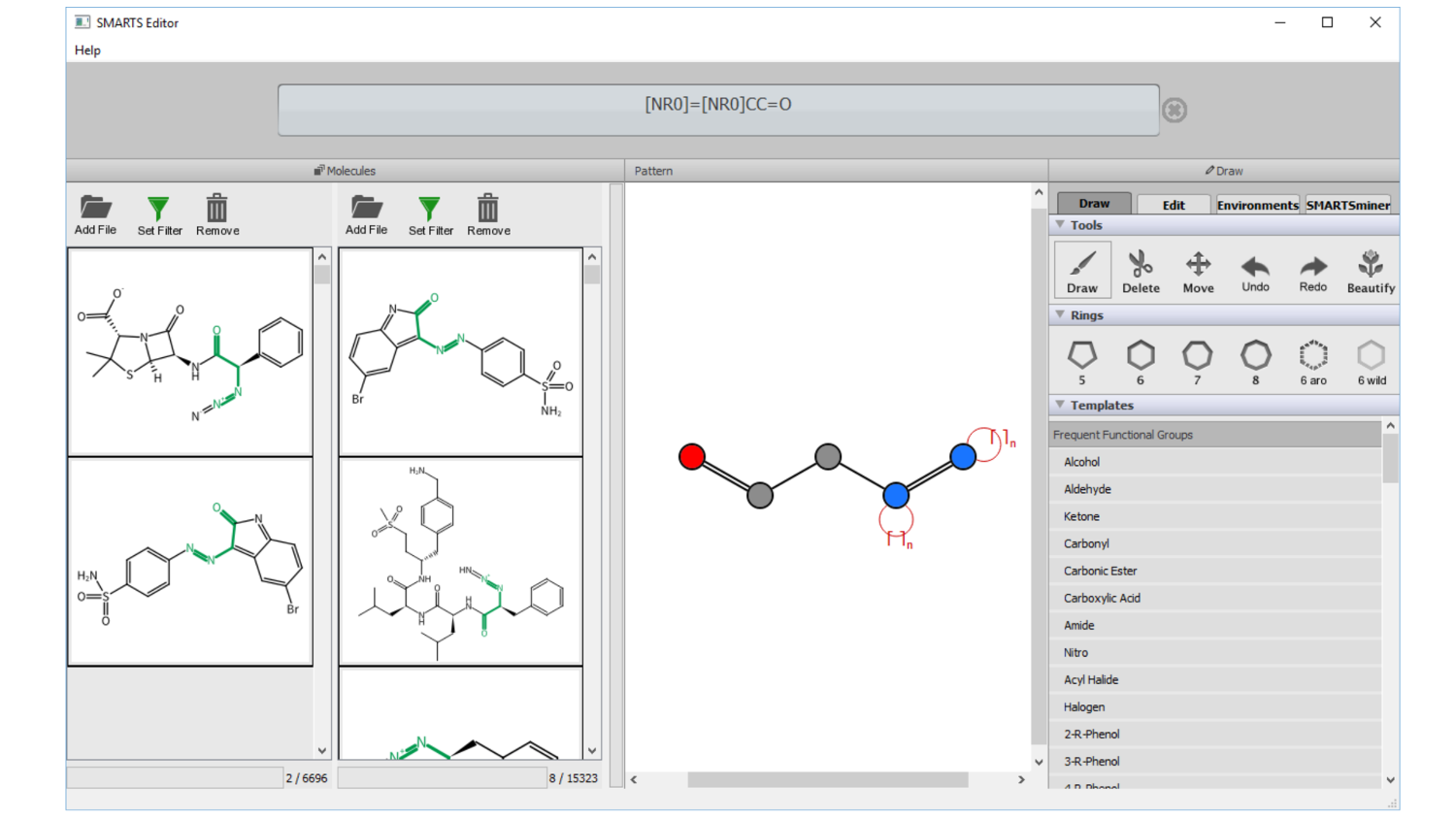
SMARTSeditor: Interactive graphical creation of SMARTS expressions
Simply draw your molecular pattern: Visualize and understand SMARTS expressions; draw scaffolds and interactively modify atomic properties; graphical navigation and specification of atomic environ-ments; on the fly pattern matching and graphical validation; fully-automatic calculation of contrast patterns separating molecule sets.

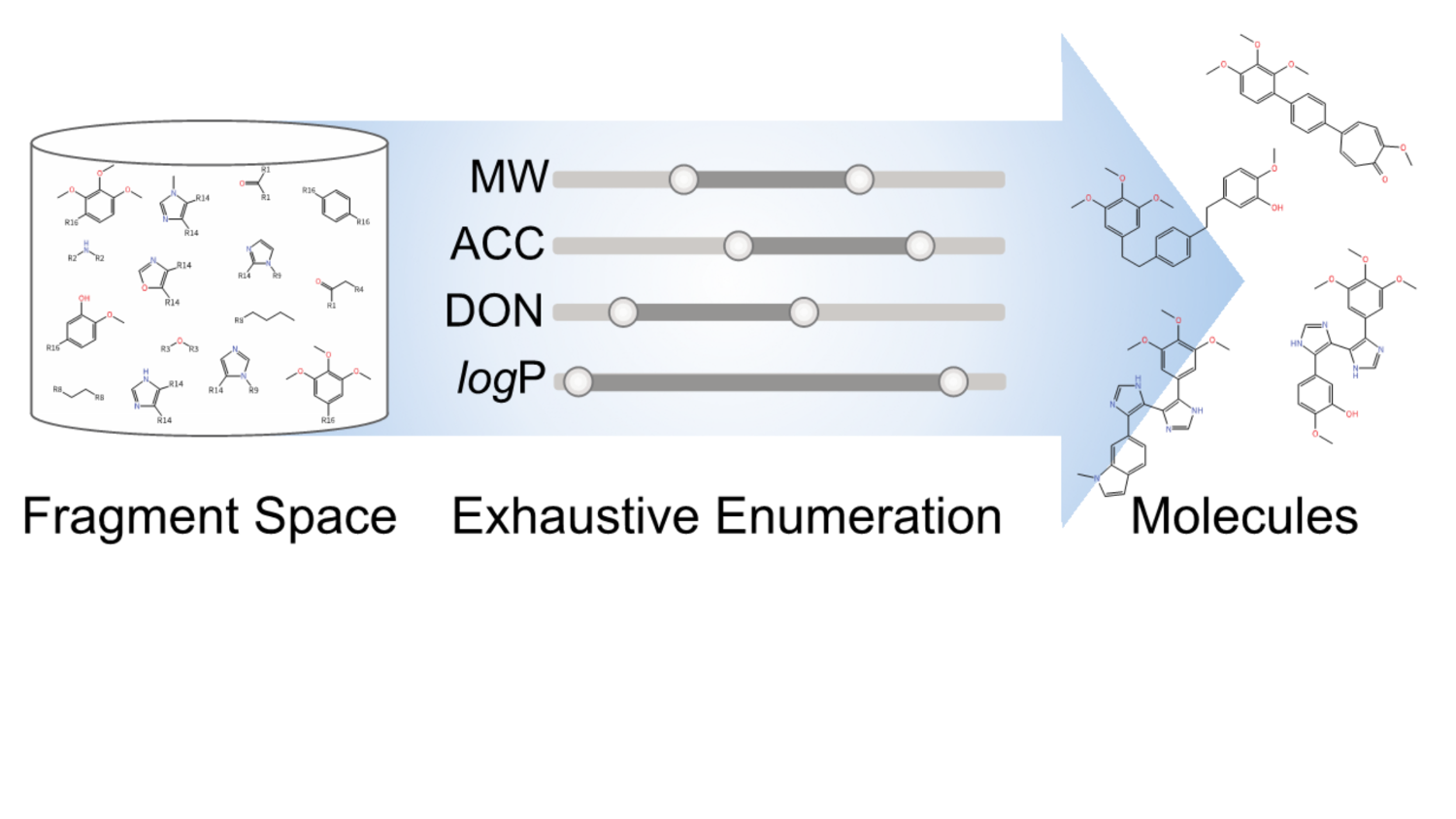
FSees: fragment space
exhaustive enumeration system
Generating large compound libraries: Enumerates a so-called fragment space under physico-chemical constraints; Ensures completeness and uniqueness umong the created compounds; Allows to limit more than ten physico-chemical properties from molecular weight via structural features to logP; Allows to combine properties to important filters (e.g. lead-like or drug-like). The basis for this method is fragment space, a fragment-oriented model of combinatorial chemistry space. It can be constructed from retrosynthetic rules or synthetic chemical reactions.

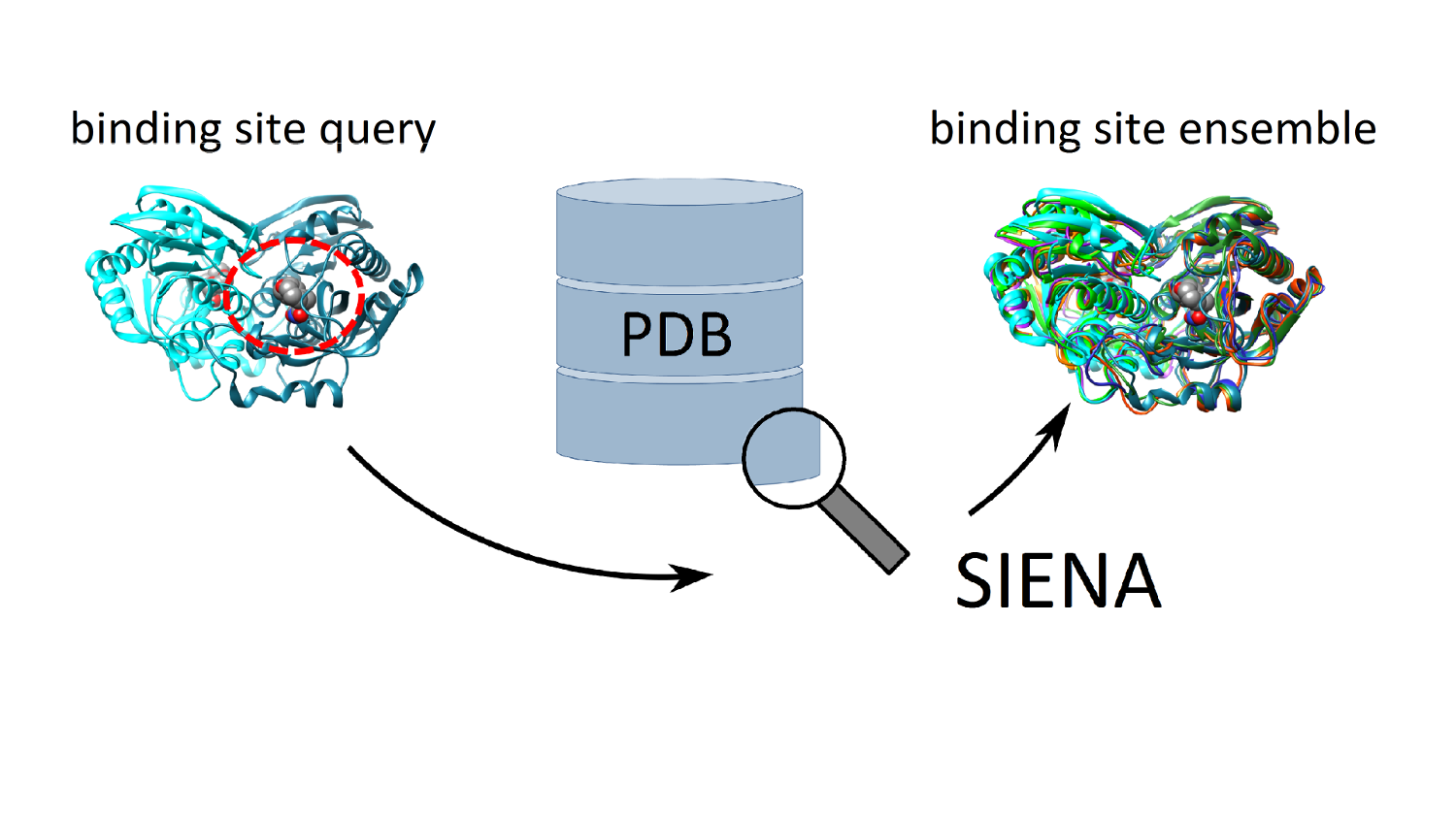
ASCONA/SIENA: Active site search and alignment
Create aligned active site ensembles: Search for sequence-similar active sites in the whole PDB; control sequence identity in the active site: find active sites in homodimeric and multimeric structures; consider structural variation upon search and alignment; create a multiple structural alignment of active sites; reduce active site collections to small ensembles with high structural variance.